Comparative in Silico Analysis of MicroRNA Binding Mechanisms in Plants and Mammals
DOI:
https://doi.org/10.37934/sijn.4.1.19aKeywords:
MicroRNA, plants, mammals, binding mechanismAbstract
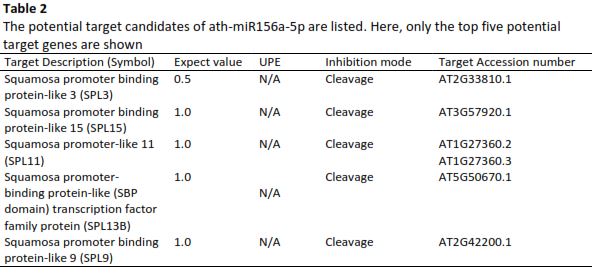
MicroRNAs (miRNAs) are essential regulators of gene expression in eukaryotes, playing critical roles in various physiological and pathological processes. While the conserved core functions of miRNAs are well-documented, significant gaps remain in understanding the contrasting mechanisms of miRNA-mediated gene regulation between plants and mammals. Specifically, the differences in target binding and regulatory outcomes are not fully elucidated, hindering a comprehensive view of miRNA functionality across biological systems. This study addresses this gap by investigating these differences using well-characterized examples of miRNAs: miR-156 and miR-172 in plants, and miR-21 and miR-29 in mammals. These miRNAs were selected because they are extensively studied and represent well-established models of miRNA function in their respective systems, enabling robust comparisons. Target prediction tools, such as psRNA Target for plants and TargetScan for mammals, were employed to analyze binding mechanisms. Bioinformatic analysis revealed multiple target genes for both plant and mammalian miRNAs. Comparisons of the best alignments indicate that plant miRNAs exhibit near-perfect complementarity with target mRNAs, leading to direct cleavage, whereas mammalian miRNAs bind with partial complementarity, primarily at the 3' UTR, resulting in translational repression or mRNA destabilization. These findings bridge the knowledge gap by providing valuable insights into the molecular interactions underlying miRNA-target gene regulation, highlighting the unique characteristics and broader implications of miRNA-mediated processes in diverse biological systems.